Modeling the growth process of neurons
Jessica Zhang’s lab has furthered its research with IGA analysis and accurately modeled the growth of a neuron.
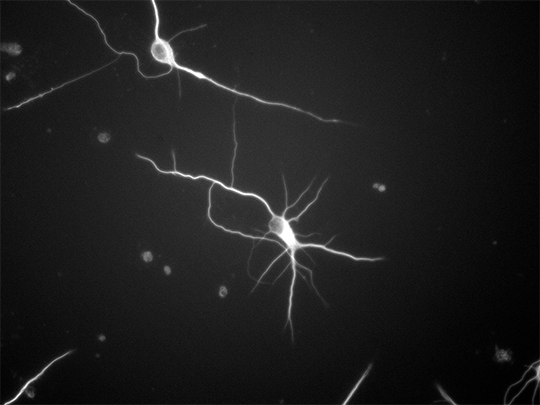
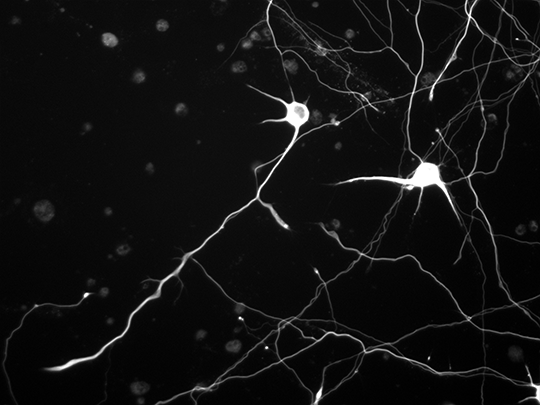
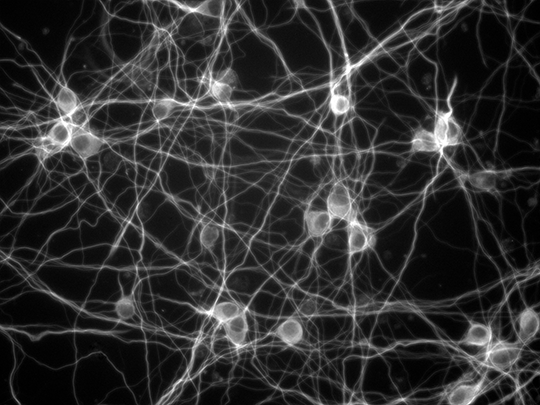
Modeling the growth cycle of a neuron is tricky, as neurons are formed for vastly different functions all over the body. However, every neuron begins its life in about the same way and goes through five distinct stages of development. A multi-disciplinary team of Carnegie Mellon University engineers led by Jessica Zhang has published a research paper outlining a new way of modeling neurons and their growth process with the goal of providing better models for studying neurodegenerative diseases, such as Alzheimer’s.
The team used a new type of isogeometric analysis (IGA) called isogeometric collocation and a phase field method. IGA is an advancement upon finite element analysis (FEA), which breaks down larger models into discrete elements that can be modeled more easily than the whole. IGA uses a method called spline geometry to produce much more accurate models whose parameters can be altered without recalculating the entire system, as is necessary in FEA. Phase field modeling is a way to simulate a continuous change in the way the model behaves when normally there would be distinct phases with different behaviors.
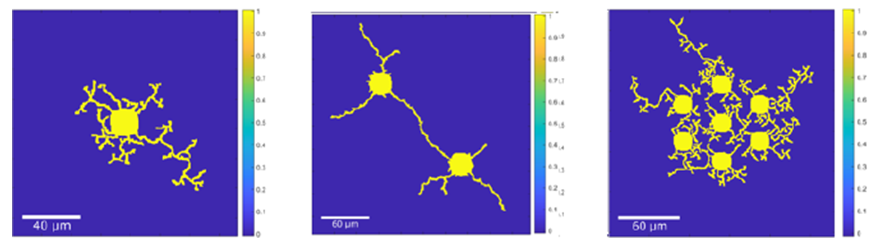
Source: College of Engineering
(A) Neuron growth model simulation result of a single neuron that exhibits growth behavior from stage 1 lamellipodia formation to stage 4 dendrite formation with observable stage 3 axon differentiation. (B) Simulation result of 2 neurons using attractive cue-guided neuron growth that shows a realistic growth pattern. (C) Simulation result of 7 neurons with all 4 growth stages that exhibit neurite interactions with each other while preventing interactions between neurites belonging to the same neuron.
The team used pictures of the growth process of neurons photographed in the Biohybrid and Organic Robotics Group (B.O.R.G) by Victoria Webster-Wood, an assistant professor of mechanical engineering, and co-advised Ph.D. student Ashlee Liao as references to build their models from.
Originally, the team tried to use the finite difference method (FDM) of solving differential equations, a very computer-friendly method that seemed like the easiest option for the job. Unfortunately, FDM did not converge very often. It would produce a result for one set of variables, but changing the starting variables slightly using the same equations would produce completely different and inaccurate results. This was not ideal as the team was trying to build a system that could be used to model all types of neuron growth patterns.
Developing the isogeometric collocation method was very difficult. The method is useful because it connects a series of equations for each stage of the growth process into one continuous equation, but because of this, each new phase adds a whole set of limitations that must be considered for the final equation.
We spent three years developing this software.
Jessical Zhang, Professor, Mechanical Engineering
“It’s a very challenging problem,” said Zhang, a professor of mechanical engineering. “We spent three years developing this software.”
The team successfully modeled the first four stages of neuron growth. “We couldn’t fully capture stage 5, the maturation stage,” said co-author Kuanren Qian, a Ph.D. student in mechanical engineering. “All types of neurons behave differently in the maturation stage based on their functions.” The parameter equations the team developed for the initial neuron growth would have to be personalized to every different type of neuron they wanted to model. “Our results, when compared to the images, yield the most realistic models,” Zhang said.
The team is currently developing a machine learning model based on the results of these growth simulations to speed up the process in order to produce more complex versions of these simulated structures. “We use machine learning to efficiently predict the neuron growth procedure,” Zhang said. The goal is that with more complex structures, researchers will be able to study different conditions that affect neuron growth and different neurodegenerative diseases, such as Alzheimer’s.
The team also consists of several collaborators, including Aishwarya Pawar, an assistant professor at Iowa State University and former Ph.D. student in Zhang’s group; Adam Feinberg, professor of biomedical engineering and materials science and engineering; and Cosmin Anitescu and Timon Rabczuk of Bauhaus University in Weimar, Germany.